SPOKE
SPOKE (Scalable Precision Medicine Knowledge Engine) is a very large network containing multiple types of biological data.
- nodes (points) within the network represent specific entities such as chemical compounds, genes, proteins, diseases, symptoms, and side effects
- edges between pairs of nodes represent known connections, for example, protein-protein binding interactions or symptoms associated with a disease
Following paths along edges in the network may reveal new connections for better understanding, treating, or preventing disease.
SPOKE is much too large and dense to comprehend visually all at once. Many analyses require intensive calculations run directly on the database. However, a Neighborhood Explorer web interface is provided for human interaction: finding and viewing specific “neighborhoods” or local subnetworks within SPOKE.
This tutorial goes step by step through some of the features of the Neighborhood Explorer. It is simply intended to illustrate these features, without promoting any particular scientific hypothesis. See also the complete documentation.
The initial appearance of the interface is something like this, with checkboxes to hide/show Options, the Sample Queries, and the node-color Legend:
– tutorial created 6/2020; results may differ as SPOKE is updated –
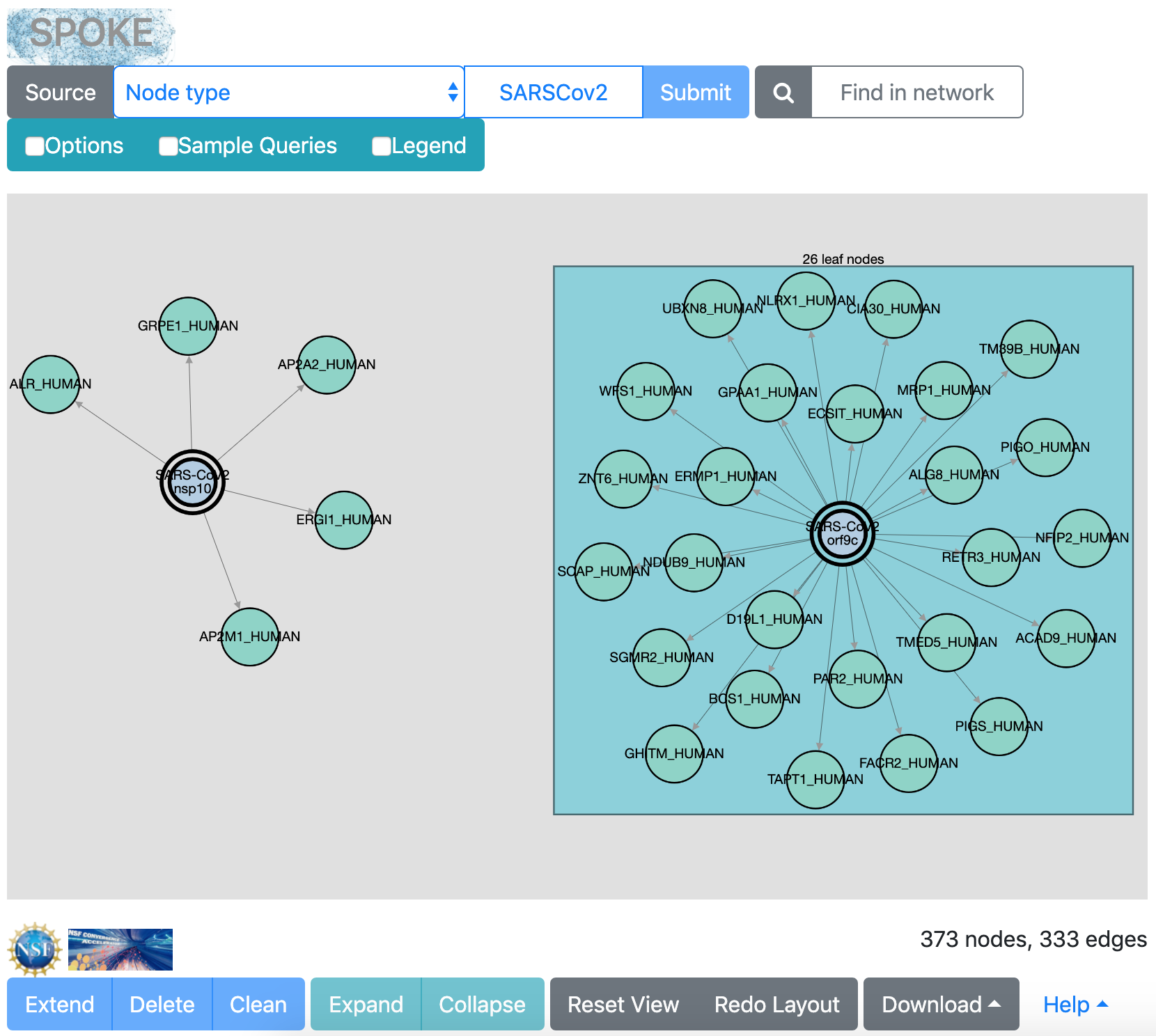
In the interface, click the sample query SarsCov-2 molecular explorer. The results may look something like this (the exact layout will vary):

This network shows interactions between human proteins and SARS-CoV-2 proteins identified in Gordon et al., Nature. 2020 Apr 30. doi: 10.1038/s41586-020-2286-9.
The experiments tested 27 coronavirus proteins, shown in the network with double outlines. All of the other nodes are the human proteins with which they interact. Boxes are drawn where many leaf nodes (those with only a single edge) surround the same central node.
Manipulation:
- scroll to zoom
- drag background to move view
- drag node to move node
- mouseover node or edge to show info
- click node or box to select
- click background to clear selection
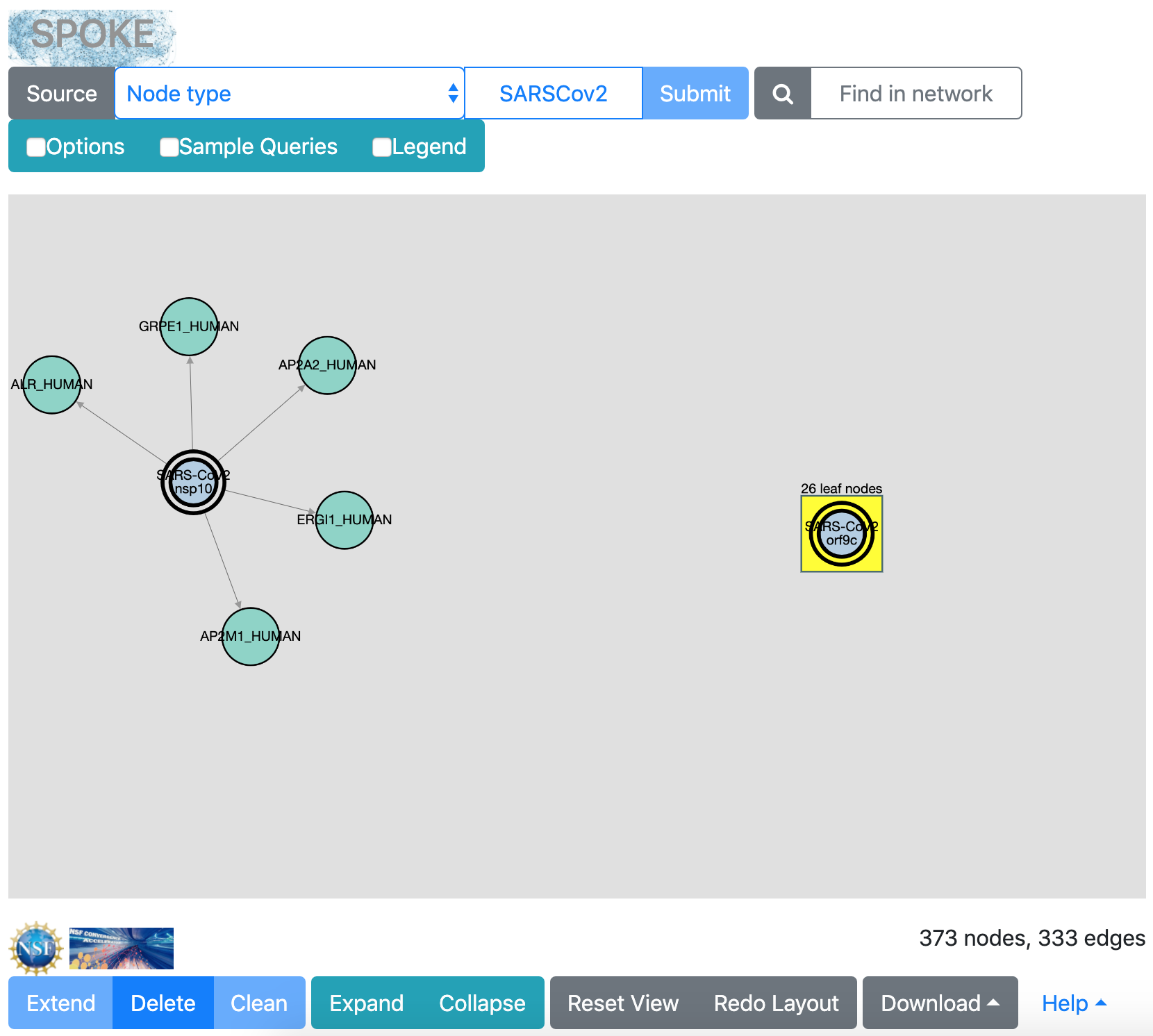
Clicking an individual node or a box “selects” it and highlights it in bright yellow. Clicking the background clears the selection. Selecting a box and then clicking the Collapse button hides the leaf nodes, leaving the box in collapsed form. This is just a way to simplify a crowded view. Clicking Expand re-shows the leaf nodes for any box that is both collapsed and selected. You can try this with any box.

|
|
Show the Options. It has three tabbed sections:
- Node and Edge Types – which node and edge types to include
- Node and Edge Attributes – filters on node and edge values
- Limits – search size limits
If the list of options does not fit in your screen, use the standard mechanism of your browser to zoom out (reduce the font size). Keep the settings from the previous search, except:
- in Node and Edge Types, un-check the Gene node type
- in Limits, increase the Maximum path length to 2
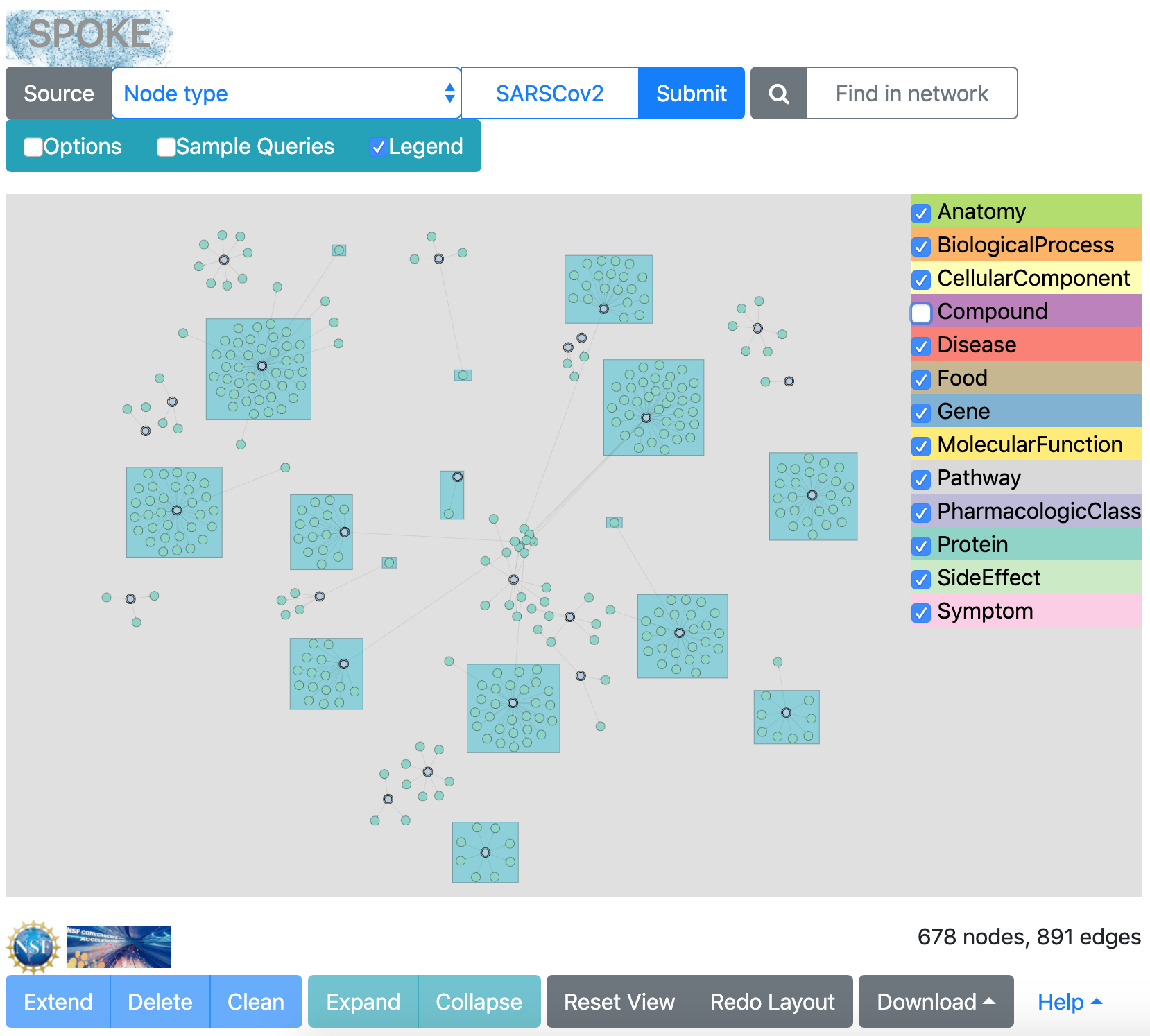
This will give the same results as the previous search, plus compounds known to bind the human proteins (Compound-binds-Protein edges). Path lengths > 1 should be used with extreme caution and only in the narrowest of searches. In this case, the coronavirus-human interactions are an extremely small dataset, and only one more edge type will be added. A too-broad search will take a very long time and even if it finishes, may give a message that the network is too large to render. There is currently no way to cancel a search in progress.
Additional edge types are still checked in the Options, but no edges to Gene nodes will be shown since that node type was excluded, nor will Compound-treats-Disease because such edges cannot be reached within 2 edges of the SARSCov2 nodes.
Click Submit. (If the button is disabled, try first re-entering “SARSCov2” in the field to the left of the button.) After the results come back, decrease the Maximum path length back to 1, then hide the Options.
The results may look something like the following. The layout will vary, and will change each time Redo Layout is clicked. The color legend includes checkboxes to hide and show specific node types; if the purple Compound nodes are hidden, the same nodes and edges as from the first search are displayed.

|
|
To facilitate viewing, remember that you can scroll to zoom, drag the background to move the view, drag individual nodes and boxes, collapse boxes, and/or click the Redo Layout button to calculate a completely new layout.
Some of the human proteins are known to bind many compounds. For example, MRP1_HUMAN (left image) is a multidrug resistance protein. Even more compounds bind SGMR1_HUMAN (Sigma 1 receptor, right image), a protein involved in a wide variety of cellular functions. The Nature paper found some of these compounds to be antiviral.
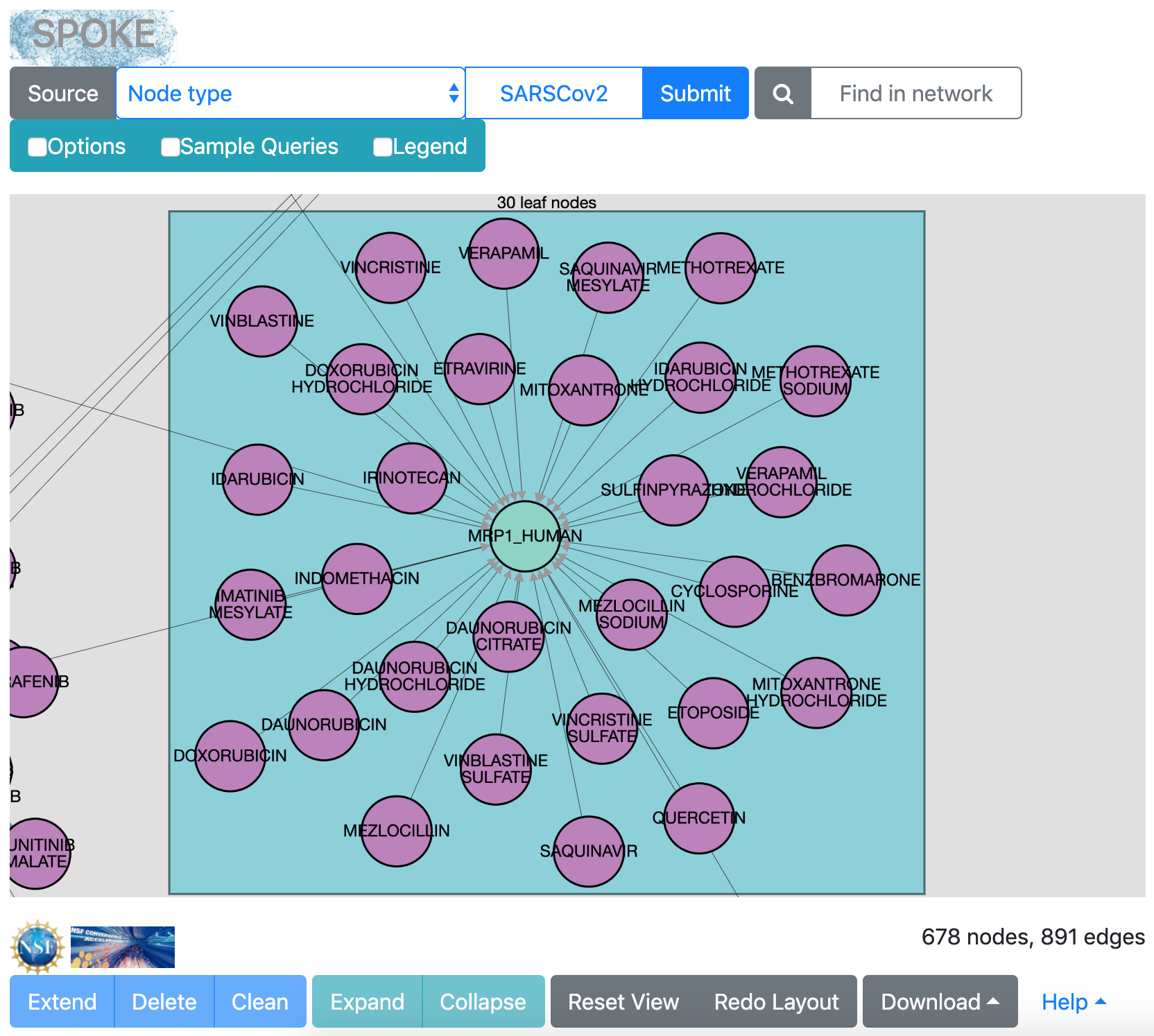
|

|
Human kinases are bunched together in the layout because many compounds are inhibitors of multiple kinases:

Next, take a closer look at the interactions of SARS-CoV-2 protein Nsp14. Enter “nsp14” in the “Find in network” search field. This will select the Nsp14 node, turning it bright yellow so that you can see where it is and zoom in on its neighborhood.

One of the proteins that Nsp14 binds is AGAL_HUMAN (the enzyme α-galactosidase), which in turn binds the compound migalastat. The Compound-treats-Disease edge type was already checked in the Options, so you can search with migalastat as the new query to see what diseases (if any) it treats. Select migalastat by clicking that node, and then click the Extend button.

Clicking Extend adds to the current network by searching with the selected node, now shown with a double border because it is the query. Eventually several human proteins that bind migalastat are added, along with one disease node, in red: Fabry disease. To make this image, the disease node was manually dragged closer to the migalastat node.
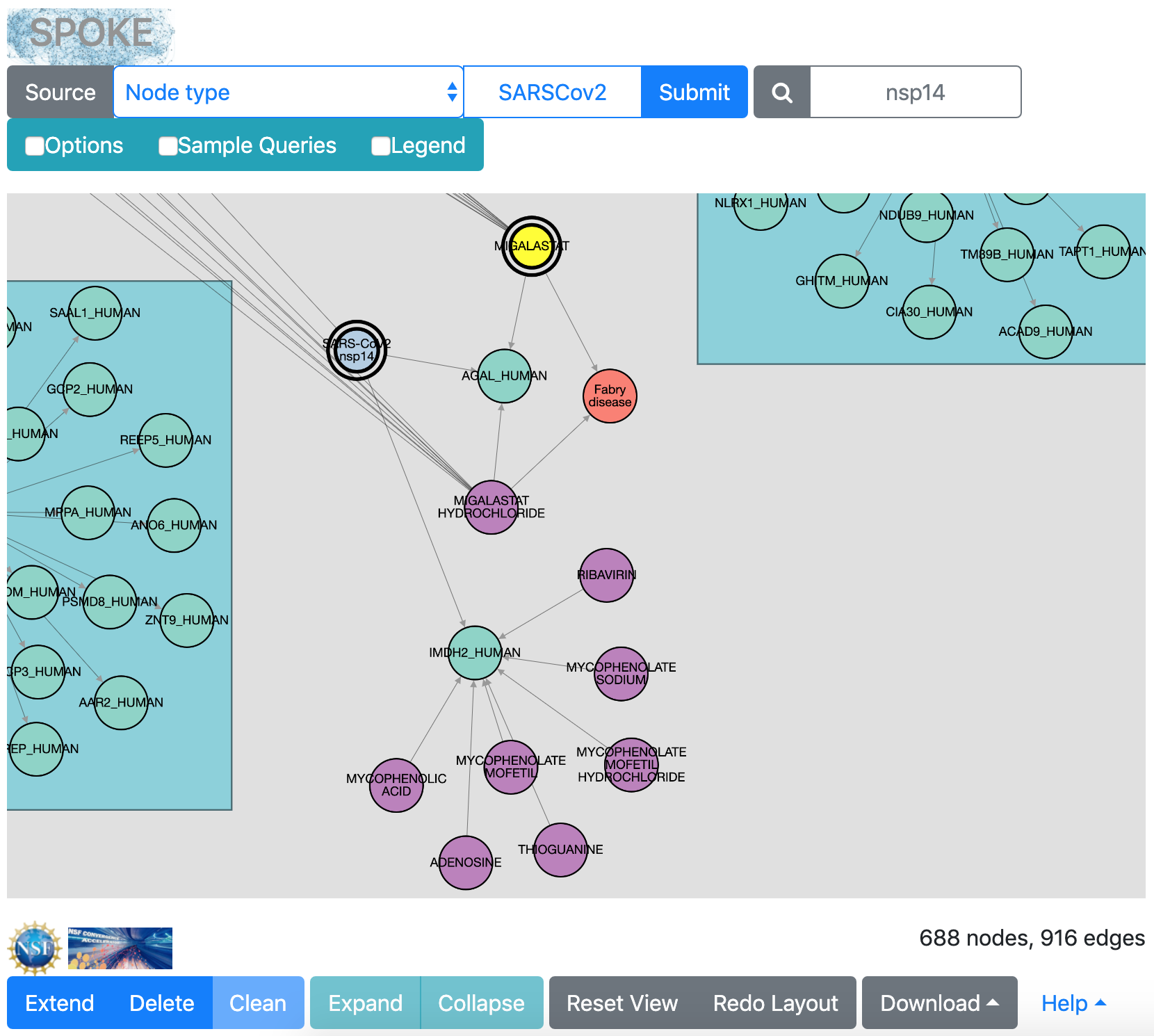
Nsp14 also interacts with IMDH2_HUMAN (inosine 5'-monophosphate dehydrogenase 2). This enzyme binds several compounds, including ribavirin. Ribavirin's mechanism of action is not completely understood, but it is part of a triple therapy under investigation for treating Covid-19 (NCT04276688).
Next we will search for investigational compounds known to bind IMDH2_HUMAN. Click that node to select it:

Show the Options again, and in the Node and Edge Attributes section, decrease the Compound max phase from 3 to 2. This will allow finding compounds that have only reached phase II clinical trials (0: pre-clinical; 1-3: the corresponding phase of clinical trials; 4: approved treatment). This setting filters Compound nodes regardless of their edges (an approved compound might still bind proteins that have nothing to do with its approved use, for example, or be used to treat diseases other than the approved indication).
Lower down, under Filter edges: The Compound-treats-disease phase setting specifically filters Compound-treats-Disease edges. Change that to 2 as well, although it will not affect the next step, which adds only Compound-binds-Protein edges.
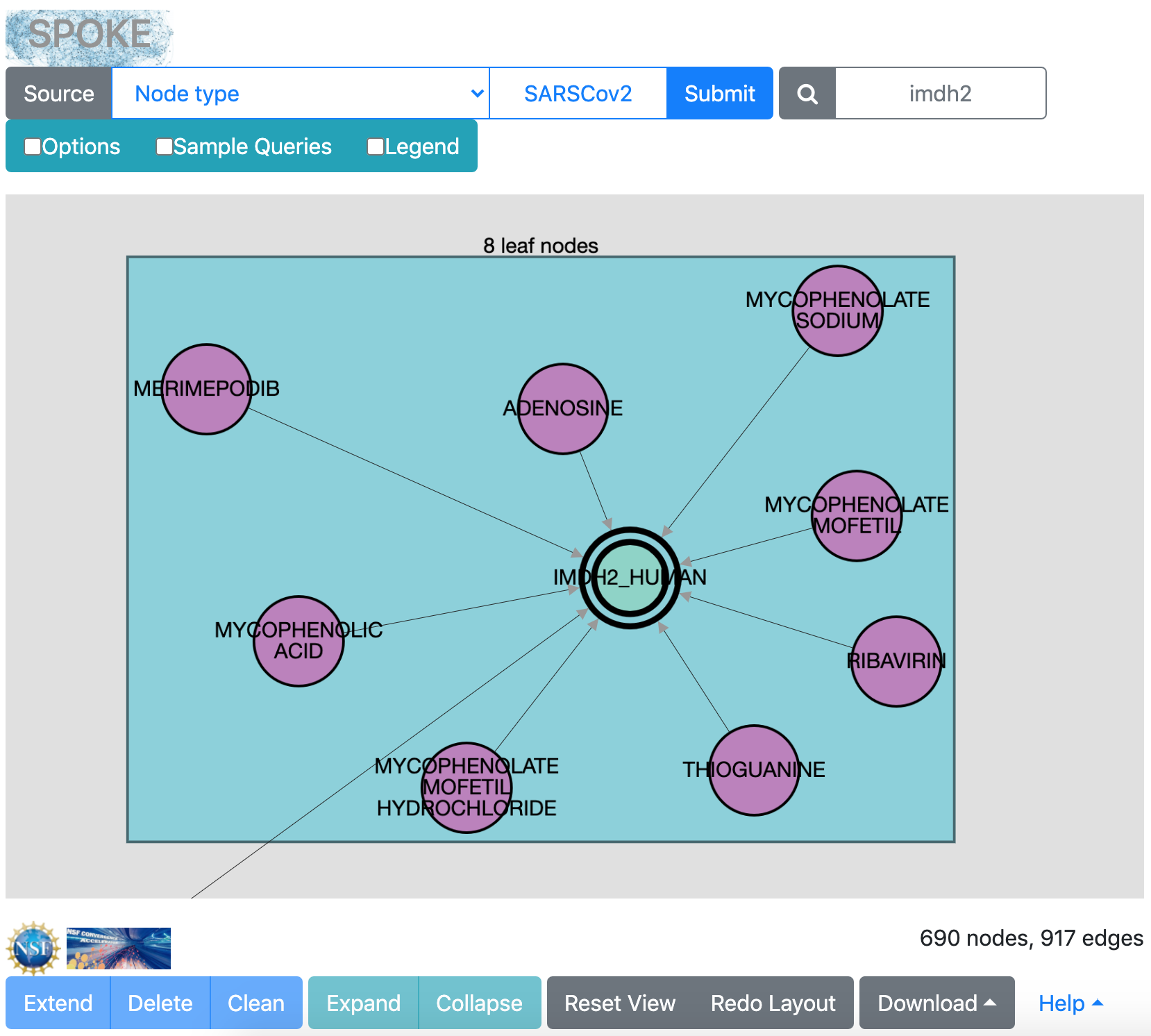
Hide the Options and click Extend to search with the selected node (IMDH2_HUMAN).
After this search, there are enough leaf nodes around IMDH2_HUMAN to
be shown in a box. If it is difficult to see what the box contains,
you can drag the box away from other parts of the network and/or click
Redo Layout. Most of the compounds were already present,
but now there is one more:
merimepodib (max_phase 2, as shown in the info
balloon from moving the cursor over that node).
Instead of running a sample query, a more common use of the Neighborhood Explorer may be to search with a specific protein, compound, disease, or gene of interest. We will end this brief tour with one example of that kind of search.
Click Node type near the top of the window to show a menu of query-type possibilities, including:
- Compound (CHEMBL identifier)
- Disease (DO identifier)
- Gene (Entrez name)
- Node type
- Organism (Species name)
- [etc.]
Choose Disease. To the left, click Source to show the Disease Ontology website. Searching that site with “migraine” gives the identifier as DOID:6364. Enter “DOID:6364” in the query field of the Neighborhood Explorer. Show the Options, clear all node types except compound, disease, and gene, and check all edge types. Hide the Options and Submit the search.
The results include other diseases that are related to migraine, compounds that may treat or are contraindicated for migraine, and genes associated with migraine. A few nodes are outside the box because they have more than one edge to migraine and thus are not technically leaf nodes. Interestingly, estradiol and levonorgestrel have both potential treatment (but only at phase II) and contraindication edges to migraine, perhaps for different groups of patients. If in Options the treatment edge filter is set to ≥3, only the contraindications are found.